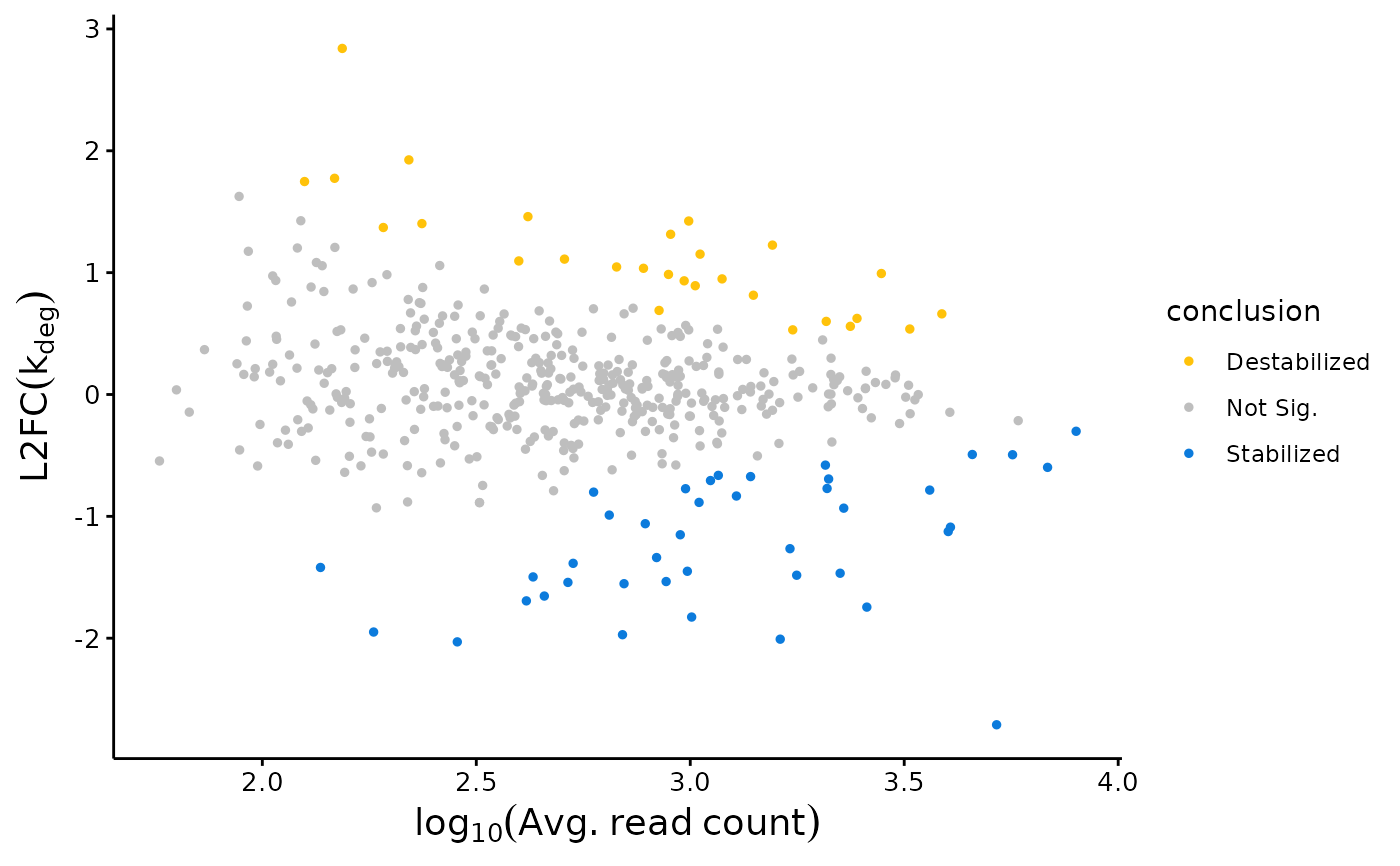
This function outputs a L2FC(kdeg) MA plot. Plots are colored according to statistical significance and the sign of L2FC(kdeg)
Usage
plotMA(
obj,
Model = c("MLE", "Hybrid", "MCMC"),
FDR = 0.05,
Exps = 2,
Exp_shape = FALSE
)Arguments
- obj
Object of class bakRFit outputted by
bakRFitfunction- Model
String identifying implementation for which you want to generate an MA plot
- FDR
False discovery rate to control at for significance assessment
- Exps
Vector of Experimental IDs to include in plot; must only contain elements within 2:(# of experimental IDs). If NULL, data for all Experimental IDs is plotted.
- Exp_shape
Logical indicating whether to use Experimental ID as factor determining point shape in volcano plot
Value
A ggplot object. Each point represents a transcript. The x-axis is log-10 transformed replicate average read counts, y-axis is the log-2 fold-change in the degradation rate constant.
Examples
# \donttest{
# Simulate data for 500 genes and 2 replicates
sim <- Simulate_bakRData(500, nreps = 2)
# Fit data with fast implementation
Fit <- bakRFit(sim$bakRData)
#> Finding reliable Features
#> Filtering out unwanted or unreliable features
#> Processing data...
#> Estimating pnew with likelihood maximization
#> Estimating unlabeled mutation rate with -s4U data
#> Estimated pnews and polds for each sample are:
#> # A tibble: 4 × 4
#> # Groups: mut [2]
#> mut reps pnew pold
#> <int> <dbl> <dbl> <dbl>
#> 1 1 1 0.0500 0.000999
#> 2 1 2 0.0500 0.000999
#> 3 2 1 0.0501 0.000999
#> 4 2 2 0.0501 0.000999
#> Estimating fraction labeled
#> Estimating per replicate uncertainties
#> Estimating read count-variance relationship
#> Averaging replicate data and regularizing estimates
#> Assessing statistical significance
#> All done! Run QC_checks() on your bakRFit object to assess the
#> quality of your data and get recommendations for next steps.
# Volcano plot
plotMA(Fit, Model = "MLE")
 # }
# }
